Utrecht Mass Spectrometry Imaging facility
This facility is a shared initiative of UMCU and UU. It is run by the Tissue Facility at the department of Pathology.
Matrix-Assisted Laser Desorption Ionization (MALDI) Mass Spectrometry Imaging (MSI) is a label free technique that is used to visualize the two-dimensional (and 3D) molecular distribution of a broad variety of molecular classes such as drugs, lipids and proteins in biological samples.
Among the several mass spectrometry ionization techniques that can be used to directly analyze tissues, MALDI has led the way in the development of biological and clinical applications for MSI.

This facility is a shared initiative of UMCU and UU. It is run by the Tissue acility at the department of Pathology.
Principle uitklapper, klik om te openen
Mass Spectrometry
Mass Spectrometry (MS) is based on the production of ions, that are subsequently separated or filtered according to mass-to-charge (m/z) ratio and detected. The resulting mass spectrum is a plot of the (relative) abundance of the produced ions as a function of the m/z ratio
Mass Spectrometry Imaging
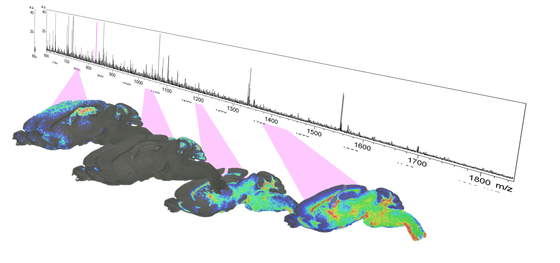
In Mass Spectrometry Imaging (MSI) spatially resolved mass spectra are being recorded by sequentially scanning the surface of a tissue sample by a laser beam, acquiring a mass spectrum from every location with a pixel size of 100-20 µm. From the data set, multiple images can be extracted, representing the spatial distribution of the analytes of interest (e.g. protein, peptide, drug or metabolite), which can be aligned with a scanned conventional staining of the same slide.
Matrix-Assisted Laser Desorption Ionization
Matrix-Assisted Laser Desorption Ionization MALDI MSI employs a laser to desorb and ionize analytes from tissues covered with a matrix that aids in the desorption/ionization process. The MSI image is then produced by scanning the sample with the laser beam as described above.
Applications uitklapper, klik om te openen
For your convenience, we have collected references to pertinent MSI literature, from sites all over the world.
| Oncology | Neuro- science | Pharma- cology |
|
|---|---|---|---|
| Proteins | Stoeckli, 2001 | Pierson, 2004 | |
| Lipids | Eberlin, 2012 | Kaya, 2017 | |
| Metabolites | Buck, 2015 | Hattori, 2010 | |
| Neurotrans- mitters | Esteve, 2016 | ||
| Glycans | Drake, 2017 | ||
| Drugs |
Equipment uitklapper, klik om te openen
Key features (provided by the manufacturer):
- Mass resolution: > 60.000 FSR (full sensitivity resolution) at 1222 m/z
- Spatial resolution: 20 μM pitch with <10 μM “zoom mode”
- Fully integrated MALDI source - switch between ESI & MALDI in seconds
- Smartbeam 3D laser with TruePixel imaging: TruePixels accurately pinpoint origin of signals and avoids oversampling errors
- SmartBeam 3D laser optics for rapid measurement: 15 pixels/second acquisition
- timsTOF provides high resolution ion mobility separation. Clear MS/MS spectra of overlapping isotope clusters
The HTX TM-Sprayer allows for uniform and reproducible matrix deposition on 2D biological samples. The HTX TM-Sprayer allows users to adjust both gas flow rate and nozzle velocity to control drying time, preventing analyte delocalization and the formation of large matrix crystals.
Slide scanner for histology and immunohistochemistry co-registered with MALDI imaging
High-end workstation with SCiLS lab 2D for data visualization, analysis and interpretation. The SCiLS Lab software implements computational methods for mining large individual MALDI imaging datasets and whole sample cohorts.

Contact uitklapper, klik om te openen
Are you interested in using this equipment or would you like to know more? You can contact the facility/Domenico Castigliego for information about the availability, tariffs, technical details and possibilities for your specific application.
T. 088 757 1518